All published articles of this journal are available on ScienceDirect.
Temporal and Spatial Expression Patterns of Bone Morphogenetic Protein 3 in Developing Zebrafish
Abstract
Bone morphogenetic proteins (BMPs) are important elements in bone biology. We herein report the expression profiles of zebrafish bmp3 (zbmp3) as demonstrated by real-time PCR and in situ hybridization. The expression of zbmp3 was highly detectable by real-time PCR from 1 day post-fertilization (1 dpf) to 2 weeks post-fertilization (2 wpf) and peaked at 1 wpf. For in situ hybridization experiments, zbmp3 was expressed in the otic vesicle at 1 dpf, 2 dpf, 3 dpf, and 5 dpf. It was also expressed in the pharyngeal arches, including the opercle, branchiostegal ray, and pectoral fins, at 2 dpf. Our results suggest that zbmp3 may play an important role in the skeletal biology of developing zebrafish.
INTRODUCTION
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-beta (TGF-beta) superfamily. Although most BMPs are positive regulators of bone biology, BMP3 is an antagonist of osteogenic BMPs [1]. The BMP3 gene is highly conserved across species, suggesting that it is evolutionarily important for development in mammals [2]. A previous study uncovered that murine Bmp3 was expressed in the cartilaginous cells of otic capsule sections using in situ hybridization [3]. However, as there have been few reports on the temporal and spatial expression profiles of Bmp3 in whole animals, we examined the expression patterns of bmp3 in developing vertebrates using a zebrafish model system.
MATERIALS AND METHODS
Extraction of Total RNA from Zebrafish Embryos and Cloning of Zebrafish bmp3 (zbmp3)
Total RNA from zebrafish embryos and larvae was extracted at selected time points using an Isogen extraction kit (Wako Co., Japan). Full-length zbmp3 (GenBank accession no. NM_001077765) was cloned into a pCR-Blunt2-TOPO vector (Invitrogen, Japan) between EcoRI sites. The primers used to amplify the zbmp3 full-length cDNA were 5’-ATGGATCGCTGTCAGCGCCTGTTTGTC CTCC-3’ (forward) and 5’-TTACCGACAGGCGCAGGAG TCCACTGTCATG-3’ (reverse). The PCR product size was 1359 bp.
Real-Time Polymerase Chain Reaction (PCR)
The primers used to amplify the zbmp3 cDNA for real-time PCR experiments were 5’-AAGGGCCATTTGGGA ACCAT-3’ (forward) and 5’-TGTGGCTGCTGTTGTG AAGA-3’ (reverse) The PCR product size was 130 bp. Real-time PCR using SSo Advanced SYBR Green supermix (BioRad, Japan) with 1-cell stage embryos (1 cell) and at 3 hours post-fertilization (hpf), 8-9 hpf, 1 day post-fertilization (dpf), 2 dpf, 3 dpf, 1 week post-fertilization (wpf), and 2 wpf were performed as described previously [4].
In Situ Hybridization
In situ hybridization was performed as earlier described [5] on 1, 2, 3, and 5 dpf embryos and larvae using full-length zbmp3. Digoxigenin-labeled RNA probes were generated using an Sp6 labeling kit (Roche, USA) according to the manufacturer’s instructions. Signals were detected with an ALP-conjugated anti-digoxigenin antibody (Roche) and visualized using 4-nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate. The reaction was stopped by replacing the substrate with several rinses of PBT (PBS plus 0.1% Tween 20). Embryos were stored in the dark at 4°C in PBS containing 0.02% sodium azide until they were photographed.
RESULTS
As shown in Fig. (1), the real-time PCR expression level of zbmp3 was very low at 1 cell, 3 hpf, and 8-9 hpf time points. However, zbmp3 expression was dramatically increased from 1 dpf to 2 wpf and peaked at 1wpf.
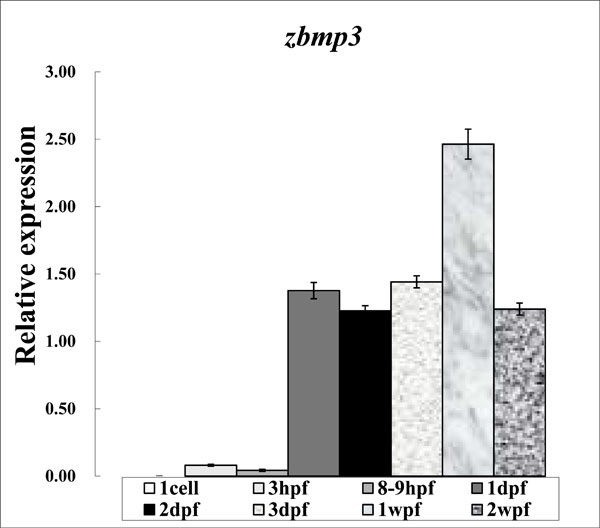
Expression study of zbmp3 by real-time PCR using samples from 1 cell, 3 hpf, 8-9 hpf, 1 dpf, 2 dpf, 3 dpf, 1 wpf, and 2 wpf time points.
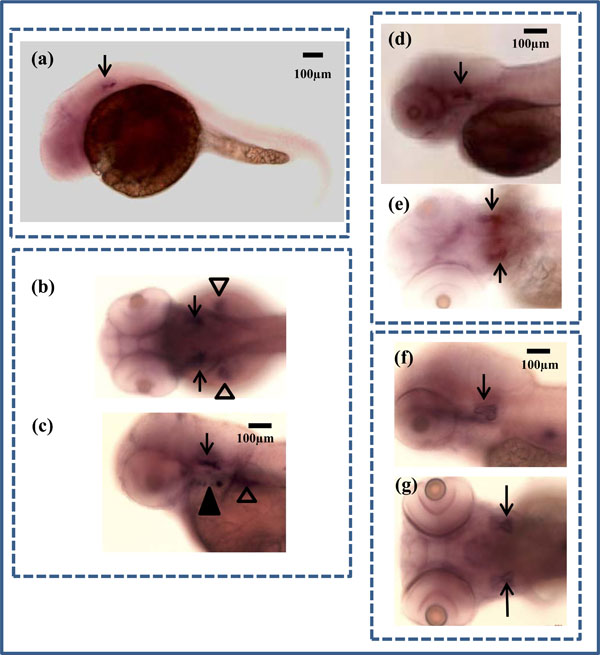
In situ hybridization of zbmp3 in developing zebrafish. (a) Lateral view of a 1 dpf zebrafish. Arrow indicates otic vesicle. (b) Dorsal view of a 2 dpf zebrafish. Arrows indicate otic vesicles and arrowheads show pectoral fins. (c) Lateral view of a 2 dpf zebrafish. Arrow indicates otic vesicle, black arrowhead shows opercle and branchiostegal ray, and white arrowhead shows pectoral fin. (d) Lateral view of a 3 dpf zebrafish. Arrow indicates otic vesicle. (e) Dorsal view of a 3 dpf zebrafish. Arrows indicate otic vesicles. (f) Lateral view of a 5 dpf zebrafish. Arrow indicates otic vesicle. (g) Dorsal view of a 5 dpf zebrafish. Arrows indicate otic vesicles.
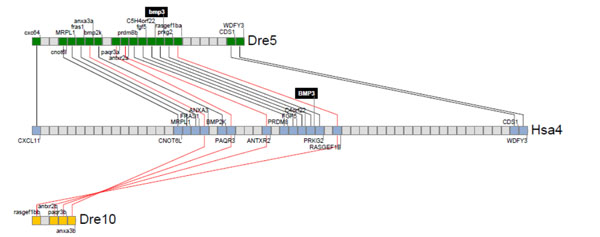
Synteny analysis for zbmp3 with human BMP3. Zebrafish and human bmp3 genomic loci were analyzed and their neighbor genes were listed in order of occurrence in zebrafish and human genomes. Shared neighbor genes were linked between zebrafish and humans to reveal evolutionary conservation of this specifc genomic locus. The bmp3 gene is indicated by a black background on zebrafish chromosome 5 and human chromosome 4.
For in situ hybridization experiments, zbmp3 was not detectable before 1 dpf, but was specifically expressed in the otic vesicle at 1 dpf (Fig. 2a; black arrows), 2 dpf (Fig. 2b, c; black arrows), 3 dpf (Fig. 2d, e; black arrows), and 5 dpf (Fig. 2f, g; black arrows). In addition, as Cheah et al. have reported previously, zbmp3 was also expressed in the opercle (op) and branchiostegal ray (bsr) (Fig. 2c; black arrowhead) and pectoral fins (Fig. 2c; white arrowhead) at 2 dpf. Each experiment was repeated at least twice. A sense (forward) probe of zbmp3 was used as a control, for which no specific signal was detected.
DISCUSSION
We observed increased and localized expression of zbmp3 at specific time points in the present zebrafish development model study. Our results suggest that bmp3 may be a key player in skeletogenesis in vertebrates.
Schoenebeck et al. have reported that zbmp3 was expressed in the pharyngeal arches and pectoral fins of zebrafish at 48-72 hpf. However, zbmp3 expression in the otic vesicle was not remarkable in their study as it was in ours (Fig. 2a-g) [6]. The reason for this disparity is not clear. Whereas Schoenebeck used an Oregon AB line [6], we adopted Cyprinidae, Rasborinae (Danio Rerio). Therefore, it is possible that a difference in genetic background may influence zbmp3 expression patterns in zebrafish lines.
The vertebrate inner ear has two major functions: 1) maintenance of body balance and 2) auditory perception. The otic vesicle plays a role in the development of ear interaction with the hindbrain [7]. Since zbmp3 was strongly expressed in the otic vesicle in zebrafish, it may influence the development of the zebrafish ear.
Runx2 is a transcriptional factor required for osteoblastic differentiation and the development of hypertrophic chondrocytes. In zebrafish, there are 2 orthologs of Runx2, runx2a and runx2b [8]. These genes were found to be expressed in the developing zebrafish skeleton, including the pharyngeal arch, op, and pectoral fins, which coincided with zbmp3 expression patterns in the present study. These results suggest a functional overlap among runx2a, runx2b, and bmp3 in zebrafish.
To our knowledge, there have been no reports on the relationship between BMP3 and rheumatologial diseases. However, other BMP family members, such as BMP4, BMP6, and Runx2, are possible biomarkers of bone metabolism in several forms of arthritis since their expression patterns are different in OA and RA groups from those in controls [9]. Based on our findings, further studies are warranted on the role of BMP3 in patients with rheumatic disease.
Gene duplication has been observed in the zebrafish genome due to an additional whole genome duplication event that is specific to the teleost fish lineage [10]. To study the conservation of Bmp3 function in vertebrates, we lastly performed a synteny analysis for zbmp3 with human BMP3 as shown in Fig. (3). We found only one zbmp3 gene, on chromosome 5, in the zebrafish genome, although a chromosomal segment duplicate on chromosome 10 that harbored a deleted zbmp3 duplicate was detected as well. Thus, one zbmp3 duplicate appears to have been lost in chromosome 10 during zebrafish genome evolution.
CONCLUSION
Our results suggest that zbmp3 plays a role in the arch skeletal system in developing zebrafish. Functional analyses using genetically modified zebrafish are needed to better understand the in vivo biological functions of zbmp3. The relationship of BMP3 with OA and RA may also require further analysis.
AUTHOR CONTRIBUTIONS
Y.N. directed this study; M.I.A. and M.M. performed experiments and drafted the article; X.H., M.H., M.M., R.W. and H.K. contributed to conception and design, interpreted the data, and revised the article. All authors approved the final version.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
Declared none.


